High-tech microscope for protein structures

A new high-tech method in microscopy holds promise for elucidating key processes of life at the molecular level: Spin-based quantum light microscopy (SQLM) combines existing analytical methods and could thus make it possible to gain a more complex picture of the biological mechanisms of proteins in the living cell. A research network at the University of Konstanz now receives 1.3 million euros in funding from the German Research Foundation (DFG) to purchase the high-tech microscope and implement SQLM in the field of structural biology and biophysics. The funding sum is provided for the purchase of the high-tech microscope in Konstanz. This technology forms the foundation for cutting-edge research projects in the life sciences.
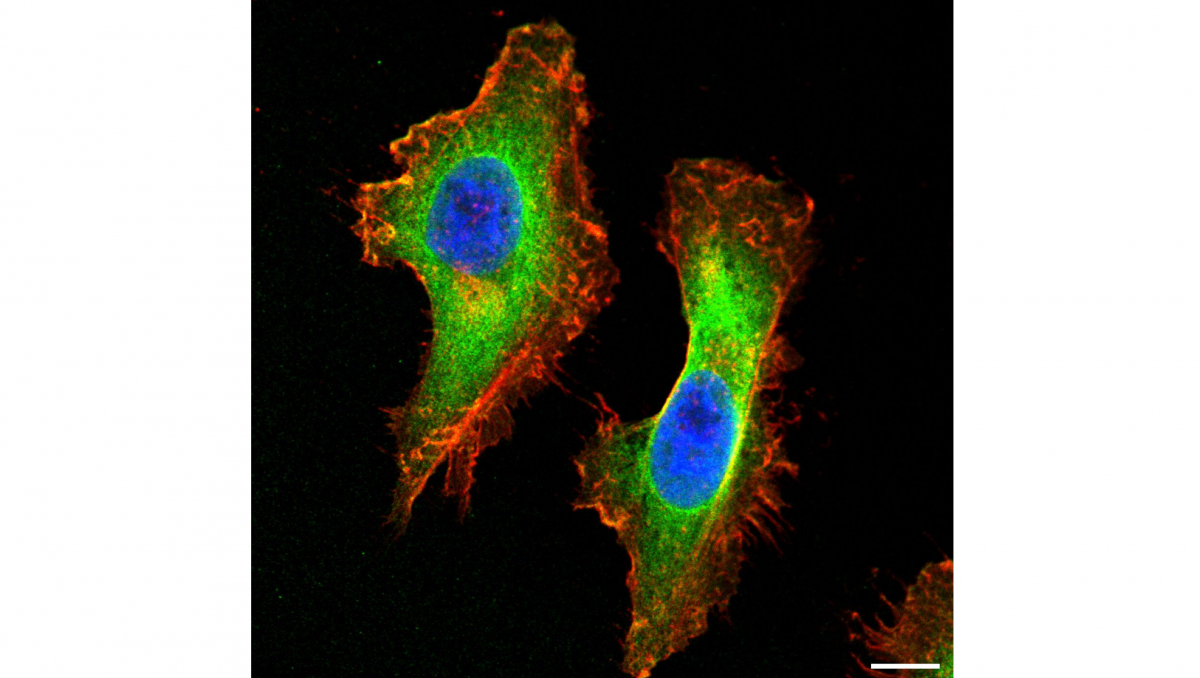
In order to decipher the secrets of life one has to look very closely. Proteins, for example, are basic building blocks of life, yet their structure and interaction with other building blocks of the body are not visible to the naked eye. Scientists typically study them in test tubes (in vitro), using advanced analytical techniques such as magnetic resonance spectroscopy and highly developed light microscopy, as is the case at the University of Konstanz. Both methods have their strengths and their specific areas of application – yet both only show part of the overall picture.
"We now want to combine both methods with each other in spin-based quantum light microscopy", says Professor Malte Drescher, spokesperson for the research network and vice rector for research at the University of Konstanz. The new method offers a combination of two perspectives on the same object and thus provides a more comprehensive overall picture of the processes.
The application of SQLM to microbiological structures is new:
© Inka Reiter"Our long-term goal is to be able to use SQLM to simultaneously observe the structure of proteins as well as their positioning, dynamics and interactions – and to do this in the living cell and at room temperature, which has thus far not been possible in this form."
Professor Malte Drescher, spokesperson for the research network and vice rector for research at the University of Konstanz
The chemist and expert in the spectroscopy of complex systems is convinced: "As a 'microscope for protein structures', spin-based quantum light microscopy has the potential to advance our understanding of proteostasis and cellular adaptation far beyond the current state of research."
Two perspectives combined: how SQLM works
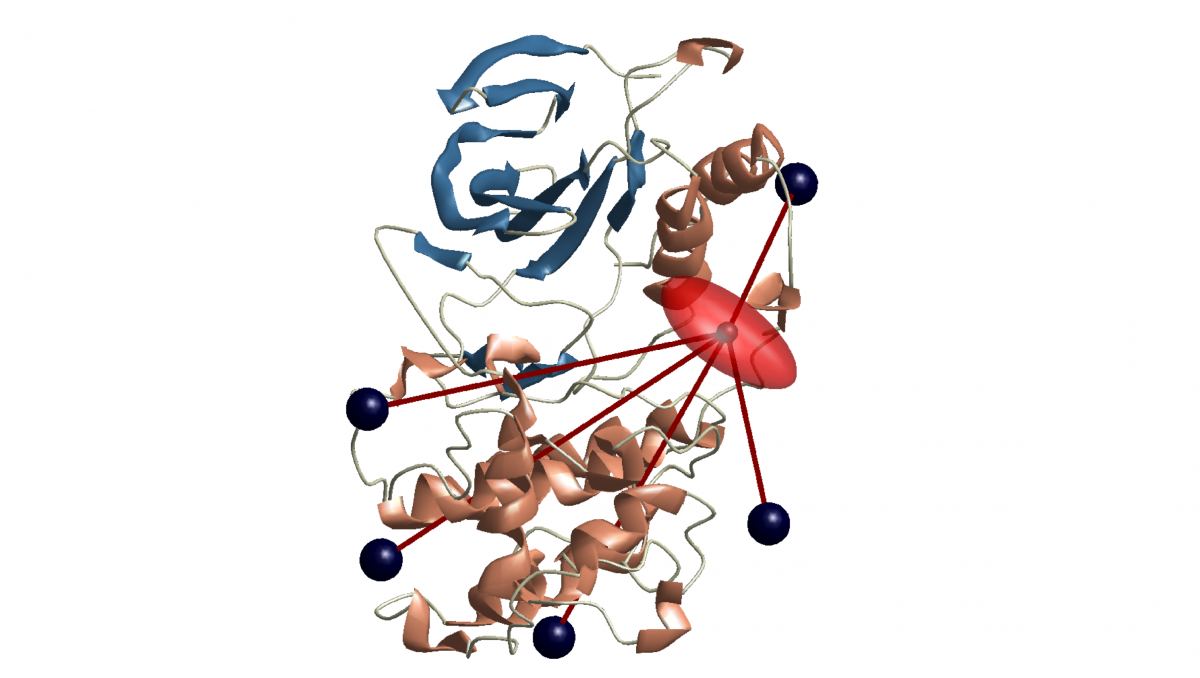
Spin-based quantum light microscopy combines light microscopy, which provides detailed information about the position of molecules, with magnetic resonance spectroscopy, which provides information about the structure of molecules (e.g. protein folding). SQLM provides two perspectives simultaneously that previously had to be obtained separately, and thus delivers an overall picture of the structure, position and dynamics of the molecules.
Put simply, SQLM uses "optically detected magnetic resonance" to gain information about molecules at the nanometre level (the thousandth part of a micrometre). We can picture the combined process as follows: We take a microscope image of a cell and, in contrast to conventional light microscopy, can now zero in on a single image point and perform magnetic resonance spectroscopy on it at the same time. The spectroscopy tells us the structure of the molecules at this image point, while light microscopy simultaneously gives us an overview of their positioning. Spin markers play a special role here: In spectroscopy, the quantum states of electron spins function as detectors that allow precise distance measurement at the nanometre level.
Transfer to scientific application
As the technology of SQLM has matured over recent years, the next step is now to apply the method in science. The research network in Konstanz sees a particularly high potential in using SQLM to analyse microbiological structures, especially proteins. Key questions the team hopes to answer with the help of SQLM include:
- How is the folding of intrinsically disordered proteins related to their position in the cell?
- How do molecular chaperones promote the correct folding of other proteins in the living cell?
- How do misfolded or damaged proteins lead to neurodegenerative diseases?
To this end, the research network in Konstanz wants to methodically advance spin-based quantum light microscopy and apply it in the field of structural biology and biophysics. The research network brings together expertise from molecular microbiology, biochemistry, spectroscopy, quantum optics, photonics and chemoinformatics, among others. In addition to the University of Konstanz, a working group from Ulm University is also involved.
Bundled competences, shared infrastructure
One milestone of the project will be the establishment of a new SQLM Core Facility at the University of Konstanz, i.e. an infrastructure and at the same time competence centre for the new SQLM method. In the spirit of the University of Konstanz's model of Core Facilities, the centre is a shared infrastructure and bundles both the technology and expertise from the participating departments.
This means: every scientist at the University of Konstanz has access to it – and external researchers can use the facility upon request. The planned Core Facility supports the research priority "Chemical Biology" at the University of Konstanz.